Ribozyme dynamics and function
Divalent ions and ribozyme catalysis
Many ribozymes (or RNA-enzymes) require ions for the chemical step of the catalysis. In this project, we investigate the molecular origin of specific ion effects in ribozyme catalysis.

In a preliminary work [1], we compared how different explicit or implicit (e.g. scaled charge, or pair-specific VdW) approaches to electronic polarization capture the interaction between divalent cations such as Mg2+ and Ca2+ and a model phosphate species, dimethylphosphate.
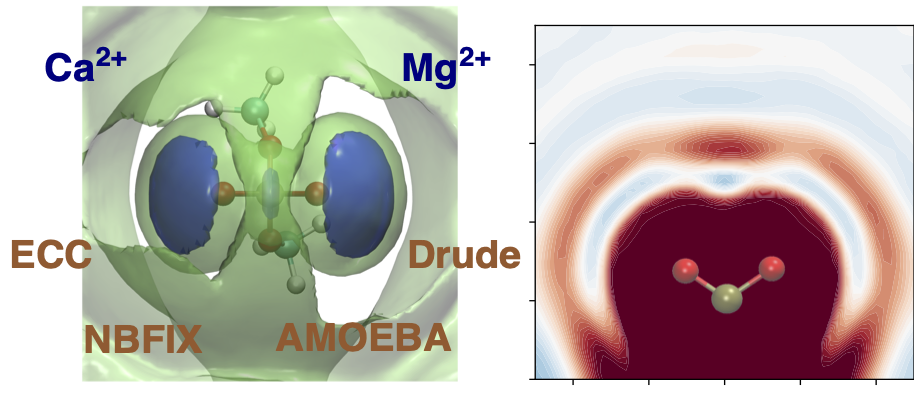
We will now combine the best ion descriptions, with enhanced sampling methologies, and dynamic mechanistic exploration with the QM/MM-MD Adaptive String Method to characterize how different cations affect both the conformation and the mechanism of the ribozyme.
This project, funded by the ANR JCJC Grant "MUSIRICAT", is carried on in collaboration with C. Clavaguéra (ICP, U. Parsis-Saclay), J. Hénin (LBT), and I. Tunon and K. Zinovjev at U. Valencia (Spain).
1. J. Puyo-Fourtine, M. Juillé, J. Hénin, C. Clavaguéra, E. Duboué-Dijon, Consistent picture of phosphate--divalent cation binding from models with implicit and explicit electronic polarization. J. Phys. Chem. B, 2022, 126, 4022-4034
Environmental modulation of the ligation/cleavage equilibrium in ribozymes
1. S. Forget, M. Juillé, E. Duboué-Dijon, G. Stirnemann, Simulation-guided conformational space exploration to assess reactive conformations of a ribozyme. J. Chem. Theory Comput., 2024, 20, 6263-6277
rRNA maturation mechanism by RNase M5 in B. subtilis
In collaboration with the experimental team led by C. Tisné at IBPC, we investigate the molecular mechanism of the final maturation step of the 5S rRNA performed by the RNase M5 in B. Subtilis. The experimental X-ray and cryo-EM structures obtained by our collaborators suggested a 2 Mg2+ ion mechanism.
This project (PhD J. Puyo) is funded by the Labex DYNAMO.
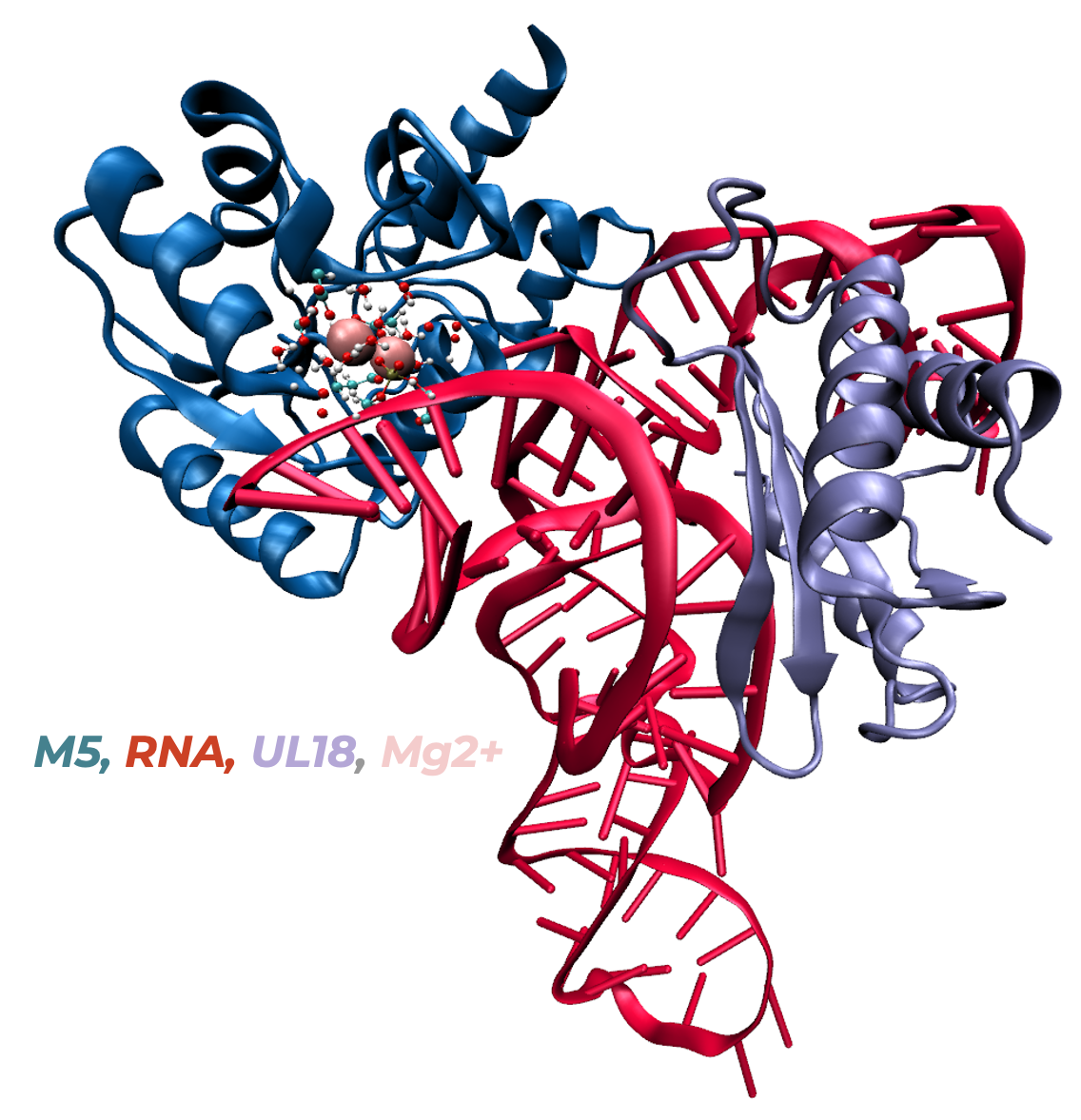
1. J. Puyo-Fourtine, L. Kantin, M. Adelkovic, C. Tisné, E. Frezza, I. Tunon, E. Duboué-Dijon, Molecular mechanism of rRNA maturation by M5: interplay between conformational flexibility and reactivity. preprint: DOI 10.26434/chemrxiv-2024-I454b
Modelling of salt solutions and salt – protein interactions
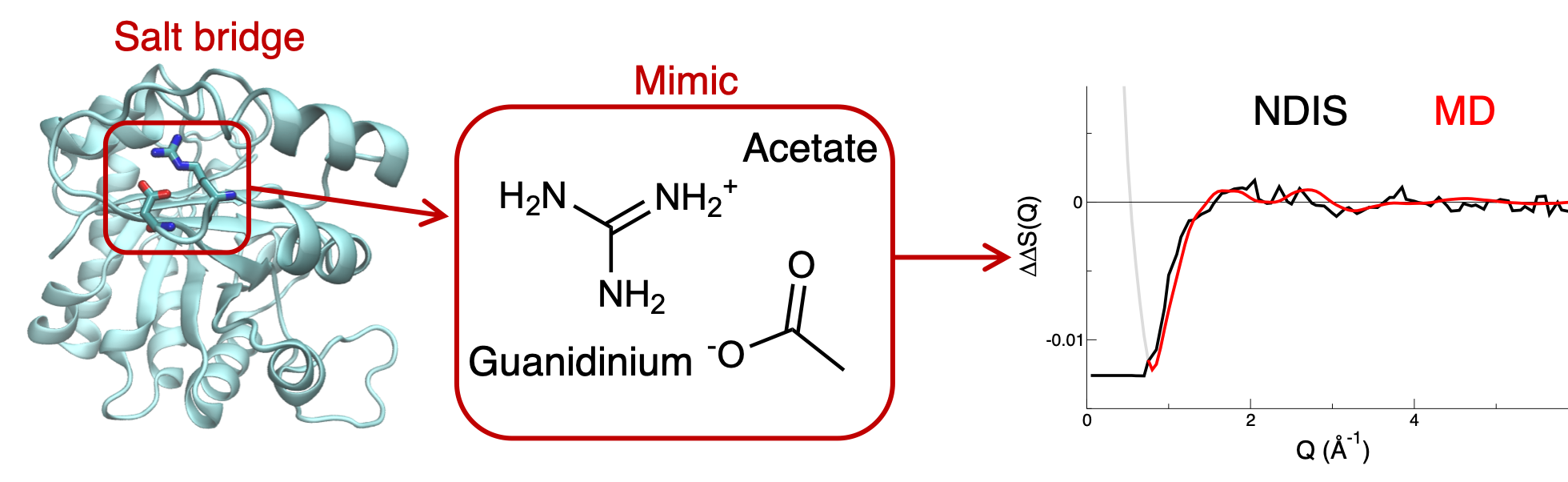
Ions are ubiquitous in biological media and are directly involved in numerous biological processes, such as calcium signalling, insulin storage or DNA compaction. Understanding how they interact with biomolecules is thus of key importance. Hence, I take part in the development of new tools to improve the simulations of salt solutions, especially in the context of biomolecular systems. We have shown that the development of scaled-charge force fields for divalent ions [1,2] and biomolecules [3], rationalized in the framework of the Electronic Continuum Correction, dramatically improves the performance of non-polarizable force field to describe ion pairing and ion-biomolecules interactions. In addition, thanks to a combination of neutron scattering experiments and simulations, we were recently able to directly characterize the strength of a model salt bridge. [4] We showed that scaled charge simulations improve the description of such salt bridge interactions compared to traditional non polarizable force fields, that strongly overestimate them. These projects are carried on in collaboration with the group of Pavel Jungwirth at the Institute of Organic Chemistry and Biochemistry, in Prague (Czech Republic),
1. T. Martinek, E. Duboué-Dijon, S. Timr, P.E. Mason, K. Baxova, H.E. Fischer, B. Schmidt, E. Pluharova and P. Jungwirth, Calcium ions in aqueous solutions: Accurate force field description aided by ab initio molecular dynamics and neutron scattering. J. Chem. Phys., 2018, 122, 10069-10076
2. E. Duboué-Dijon*, P.E. Mason, H.E. Fischer, P. Jungwirth*, Hydration and ion pairing in aqueous Mg2+ and Zn2+ solutions: force field description aided by neutron scattering experiments and ab initio molecular dynamics simulations. J. Phys. Chem. B, 2017, 122, 3296-3306
3. E. Duboué-Dijon, P. Delcroix, H; Martinez-Seara, J. Hladilkova, P. Coufal, T. Krizek and P. Jungwirth, Binding of divalent cations to insulin: Capillary electrophoresis and molecular simulations, J. Phys. Chem. B, 2018, 122, 5640-5648
4. P.E. Mason, P. Jungwirth, E. Duboué-Dijon*, Quantifying the Strength of a Salt Bridge by Neutron Scattering and Molecular Dynamics. J. Phys. Chem. Lett., 2019, 10, 3254-3259
5. E. Duboué-Dijon, M. Javanainen, P. Delcroix, P. Jungwirth, H. Martinez-Seara, A practical guide to biologically relevant molecular simulations with charge scaling for electronic polarization, J. Chem. Phys., 2020, 153, 050901
6. D. Mendes de Oliveira, S.R. Zukowski, V. Palivec, J. Hénin, H. Martinez-Seara, D. Ben Amotz, P. Jungwirth, E. Duboué-Dijon, Binding of Divalent Cations to Acetate: Molecular Simulations Guided by Raman Spectroscopy, Phys. Chem. Chem. Phys., 2020, 22, 24014-24027
7. P.E. Mason, T. Martinek, B. Fabian, M. Vazdar, P. Jungwirth, O. Tichacek, E. Duboué-Dijon, H. Matinez-Seara, Hydration of biologically relevant tetramethylammonium cation by neutron scattering and molecular dynamics. Phys. Chem. Chem. Phys., 2024, 26, 3208-3218
Biomolecular Hydration Dynamics
In the line of my PhD work, performed in Damien Laage's group at the École Normale Supérieure, I keep a strong interest in biomolecular hydration. The challenge we are trying to adress is to rationalize at the molecular level the origin of the perturbation in water dynamics next to biological solutes, and to understand how this could affect key biological processes such as DNA-protein binding. We previously characterized and mapped [1] the heterogeneity in a model DNA hydration shell dynamics and showed that it was of two kinds: the static heterogeneity originates both from the heterogeneity in the chemical properties and topography of the DNA surface, and the dynamical heterogeneity results from the modulation of hydration dynamics by DNA conformational fluctuations, notably in the minor groove. Further work is in progress to examine and rationalize the dependence of such heterogeneity on DNA sequence and extend it to RNA.
Collaborators: Damien Laage (École Normale Supérieure, PSL University, Paris) and Elisa Frezza (Université de Paris)

1. E. Duboué-Dijon, A.C. Fogarty, J.T. Hynes, D. Laage, Dynamical disorder in the DNA hydration shell, J. Am. Chem. Soc., 2016, 138, 7610-7620
2. E. Frezza, D. Laage, E. Duboué-Dijon*, Molecular origin of distinct hydration dynamics in double helical DNA and RNA sequences, J. Phys. Chem. Lett., 2024, 15, 4351-4358
Miscellaneous
In collaboration with my colleague Jérôme Hénin at LBT, we recently [1] explored subtilities in (absolute) binding free energy calculations, suggesting a theoretical framework that should allow non expert practitionners to avoid common mistakes. We also attempt to clarify how to properly treat symmetric systems.
1. E. Duboué-Dijon, J. Hénin, Building intuition for binding free energy calculs: Bound state definition, restraints, and symmetry, J. Chem. Phys., 2021, 154, 204101
